Unable to Match Data in this Source with Reference
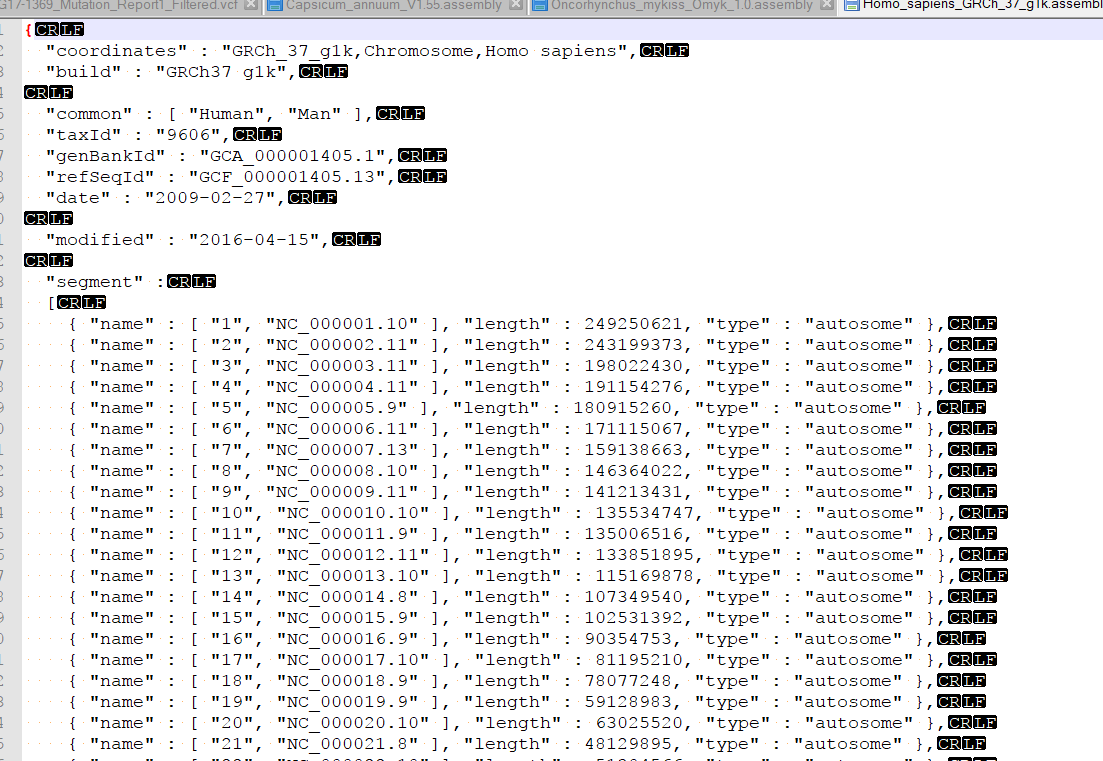
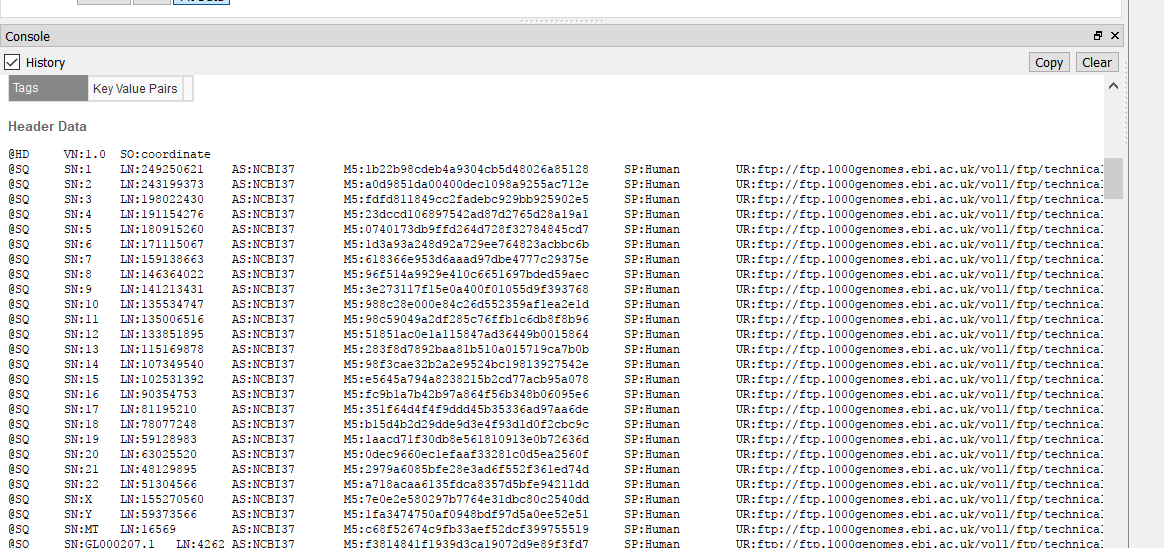
I am examining Illumina data aligned to a scaffold-level genome (30,000 contigs). I can browse these BAMs in other genome viewers (like IGV), but my preference is GenomeBrowse for numerous reasons. I have converted the very same genome fasta to GB's format, but unlike the dozen or so other species I have examined using GenomeBrowse, the indexed BAM files will not load. I receive "Unable to match data in this source with an existing reference sequence. Reference sequence must have matching chromosome names and lengths." I have validated that the names are the same (even though it's the exact same file) - they are based on NCBI accessions, and that lengths correlate to the correct contig. Is this a known problem?
Thank you in advance, -Brian