| 1 | initial version |
Hi Brian,
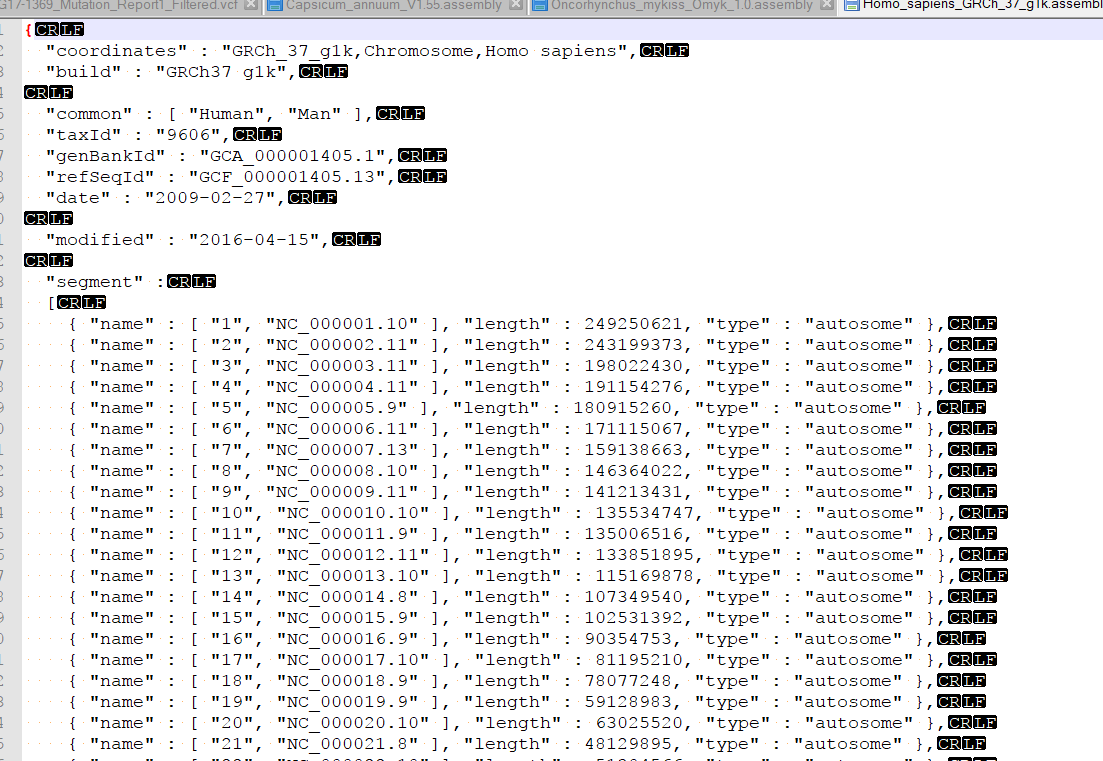
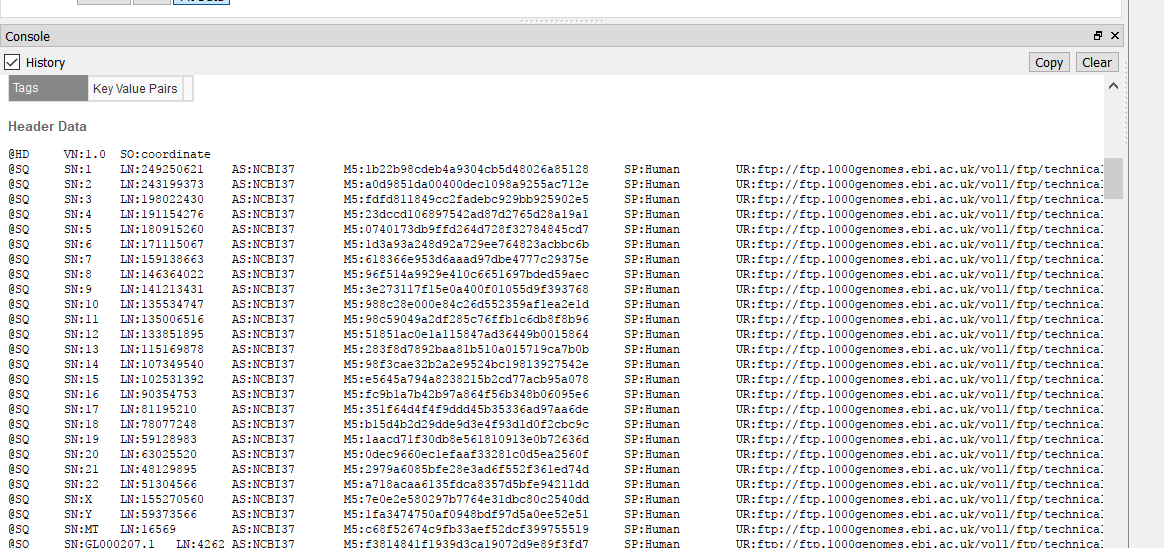
This sounds like there is a discrepancy between the headers in your BAM file and the names of the scaffolds in the assembly file that is created when converting your genome fasta in GB. You could check by plotting the BAM in GB and under the console window, examining the coordinates in the BAM file.
Then compare these coordinates to what is found in the assembly file. This is typically found in your user data folder. To access this folder from GenomeBrowse, you can click tools > Open Folder > Annotations folder. Then back-up one directory to find the Assemblies folder. you can open the assemblies file in a text editor and change the order of the chromosomes.