I would like to convert 23andme data but am stopped by step below?
"A local copy of dbSNP was not provided to query for rsID." How do I reference this?
"A local copy of dbSNP was not provided to query for rsID." How do I reference this?
Hi Janet,
That message just means that you need a local copy of a dbSNP source downloaded for the import to work correctly. You can do this by going to Tools > Manage Data Source and selecting a dbSNP track from the Public Annotations repository then click Download at the bottom of the window.
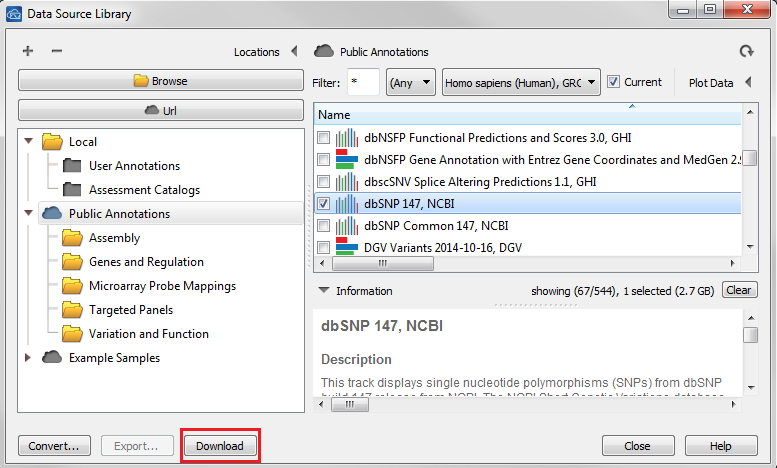
Once the download is finished you may need to hit refresh in the upper right corner of the dialog for the track to appear in your Local annotation folder. Once you can see the track in the Local folder then proceed to the Convert Wizard and add your 23andMe data, it should correctly pick-up the newly downloaded SNP track automatically.
Let me know if you have any further issues.
Thanks, Jami... Golden Helix, Inc.
Questions should be tagged FeatureRequest for asking about a non-existing feature or proposing a new idea, GeneralInquiry for general questions about GenomeBrowse or directions on how to do something, or RanIntoProblem if you want to report an issue or had difficulty getting to an expected result.
Asked: 2016-10-10 14:04:15 -0600
Seen: 3,771 times
Last updated: Oct 11 '16