Hi Whit,
I am sorry you are having issues visualizing your data in GenomeBrowse.
With mitochondrial data on the GRCh37 genome you will need to make sure you have the correct reference assembly specified in the drop down at the top of the GenomeBrowse window that matches the chromosome naming convention for this region. In particular, for the hg19 version of the assembly chrM is used to define this region but for the g1k version of the assembly chrMT is used.
If you click on the name of your BAM file in the GenomeBrowse plot and then scroll to the bottom of the Console window you will see how this region is identified in your BAM file. Should look similar to the below screenshot.
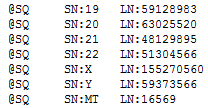
So you will want to make sure and select the appropriate assembly for your data. If that does not work to fix the visibility can you try deleting the index file (BAI) that should be in the same directory as the BAM file? GenomeBrowse will then try and recompute the file if there are issues with the data you should get some error messages that may help diagnose what is going wrong. Screenshots of any error messages would be most useful.
The read depth of the data should not cause this issues, when the number of reads exceeds what can be rendered you will still see those that fit you will just get and error message saying not all reads could be drawn.
Let me know how I can help you further.
Thanks,
Jami...


According to the company that produced the above-mentioned mtDNA BAM files, the read depth is about 2000X. Is that too much to be displayed in the GH browser? Could that be causing the problem?
Whit Athey ( 2016-05-12 12:31:55 -0600 )edit