 | 1 | initial version |
Hi Shaun,
To add a new genome assembly to GenomeBrowse requires the nucleotide sequence in FASTA format. Once you have the appropriate files you can go to Add then select Convert from the lower left corner of the Data Source Library.
This will open up the Convert Wizard so you can add in your custom data source. You can find a description of converting the FASTA file(s) in our manual at the following link. http://doc.goldenhelix.com/GenomeBrowse/latest/gbmanual/convertsourcewizard.html#converting-a-fasta-file
Once the file is converted to a usable format you can select the assembly from dropdown list.
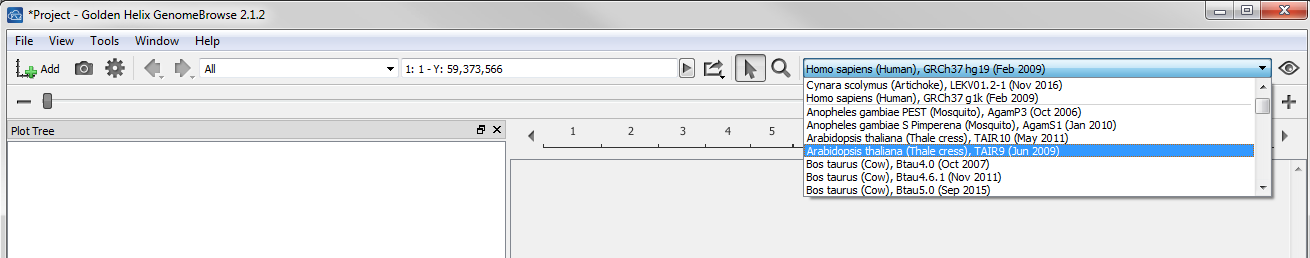
Then select the Add button to choose the new file from your Local annotations folder.
Let me know if you have any further questions.
Thanks, Jami...